Worked Example - NRMP: Dominant Frequency¶
This example demonstrates the application of dominant frequency analysis to a synthetic spatio-temporal dataset. The dataset comprises a time series of 2D images, representing the evolution of wave patterns over both space and time. By analysing the dominant frequencies at each spatial location, we can gain insights into the spatial distribution of oscillatory behaviour and identify potential wave modes.
Analysis and Figure
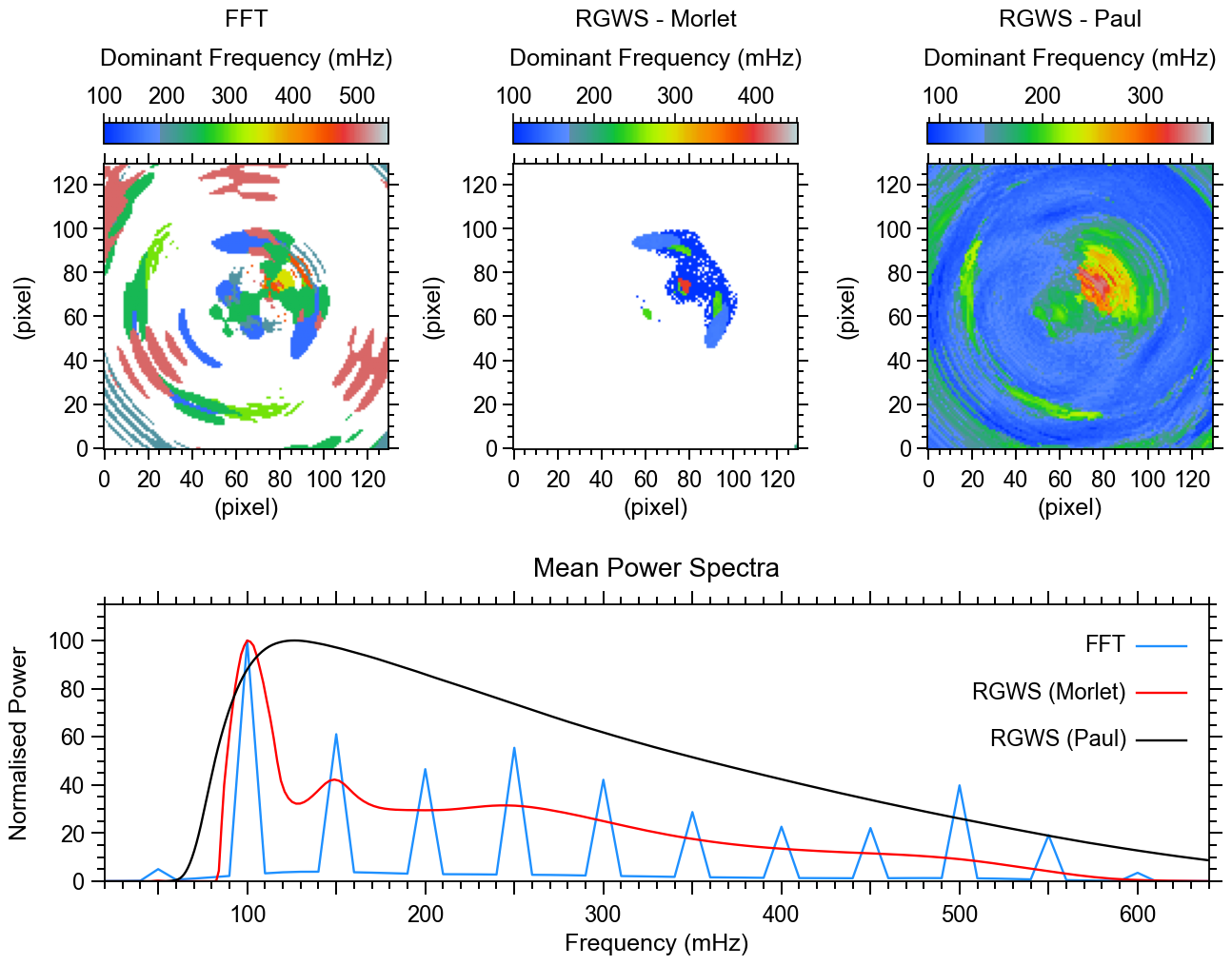
The figure below shows the dominant frequency maps calculated using different spectral analysis methods. The maps reveal the spatial distribution of the most prominent oscillation frequencies in the dataset.
Methods used:
- Fast Fourier Transform (FFT)
- Refined Global Wavelet Spectrum (RGWS) with Morlet wavelet
- Refined Global Wavelet Spectrum (RGWS) with Paul wavelet
WaLSAtools version: 1.0
These particular analyses generate the figure below (Figure 4 in Nature Reviews Methods Primers; copyrighted). For a full description of the datasets and the analyses performed, see the associated article. See the source code at the bottom of this page (or here on Github) for a complete analyses and the plotting routines used to generate this figure.

Figure Caption: Dominant frequency maps and mean power spectra. Top row: Dominant frequency maps derived using FFT (left), Morlet-based RGWS (middle), and Paul-based RGWS (right). Bottom panel: Normalized mean power spectra for FFT (blue), Morlet-based RGWS (red), and Paul-based RGWS (black).
Important Notes on Interpreting the Dominant Frequencies
The dominant frequency at each pixel corresponds to the frequency with the highest spectral power. While a linear detrending and Tukey apodization (with 0.1 tapering fraction) have been applied to reduce long-term trends and edge effects, the lowest frequency bin may still appear dominant — especially in regions without strong higher-frequency oscillations.
This is a common outcome in time series with broad, low-frequency variability, where power is not concentrated at narrow peaks, or limited by frequency resolution. However, this does not necessarily reflect meaningful or coherent low-frequency wave activity.
In the figure, white regions indicate pixels where the dominant frequency falls into the lowest frequency bin, which is explicitly mapped to white in the color table (see line 23 of the plotting routine). These are only visible in the FFT and Morlet-based RGWS maps.
The Paul-based RGWS map, on the other hand, does not show white regions. This is because, for this method, the dominant frequencies tend to fall slightly above the lowest bin (around 100-120 mHz), even though the method includes lower frequencies. This behavior reflects the fundamental differences among the analysis techniques used:
📌 FFT (Fast Fourier Transform)
◍ Provides relatively high frequency resolution at all frequencies, with fixed, global basis functions.
◍ Sensitive to any persistent oscillatory signal, but often returns low-frequency peaks if stronger high-frequency signals are absent.
◍ In the averaged power spectrum, the dominant peak lies at 100 mHz — the lowest available frequency bin.
📌 Morlet-based RGWS
◍ Uses the Morlet wavelet, which offers good frequency resolution but less precise time localization, particularly well-suited to capturing persistent oscillations with quasi-stationary frequencies.
◍ As with all RGWS methods, the frequency resolution is non-uniform, leading to narrower and more pronounced peaks at lower frequencies, and broader, smoother features at higher frequencies in the power spectrum.
◍ This causes the spectrum to mirror FFT’s tendency to favor the lowest bin as dominant when no strong high-frequency signals are present, but with less detail in the dominant frequency map, compared to FFT, due to smoothing at high frequencies.
📌 Paul-based RGWS
◍ Employs the Paul wavelet, which has excellent time localization but poorer frequency resolution, particularly at low frequencies. This causes the lowest frequency bin to appear more diffuse, with power spread across a broader range, making sharp low-frequency peaks less distinguishable.
◍ It responds strongly to short-lived or localized features in the data, but tends to underrepresent broad, slowly varying components. As a result, power in the lowest frequency bin is often weakened or distributed, making it less likely to be selected as dominant.
◍ Consequently, the mean power spectrum peaks around 120 mHz, and white regions do not appear in the dominant frequency map, since the 100 mHz bin is rarely the strongest in this method.
These differences emphasize that dominant frequency maps depend strongly on the method used, and should always be interpreted in tandem with the full power spectra (see bottom panel of Figure 4) and with awareness of each method's strengths and limitations.
Source code
© 2025 WaLSA Team - Shahin Jafarzadeh et al.
This notebook is part of the WaLSAtools package (v1.0), provided under the Apache License, Version 2.0.
You may use, modify, and distribute this notebook and its contents under the terms of the license.
Important Note on Figures: Figures generated using this notebook that are identical to or derivative of those published in:
Jafarzadeh, S., Jess, D. B., Stangalini, M. et al. 2025, Nature Reviews Methods Primers, 5, 21
are copyrighted by Nature Reviews Methods Primers. Any reuse of such figures requires explicit permission from the journal.
Free access to a view-only version: https://WaLSA.tools/nrmp
Supplementary Information: https://WaLSA.tools/nrmp-si
Figures that are newly created, modified, or unrelated to the published article may be used under the terms of the Apache License.
Disclaimer: This notebook and its code are provided "as is", without warranty of any kind, express or implied. Refer to the license for more details.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 | |
Processing FFT for a 3D cube with format 'txy' and shape (200, 130, 130).
Calculating Dominant frequencies and/or averaged power spectrum (FFT) ....
Progress: 100.00%
Analysis completed.
Processing Wavelet for a 3D cube with format 'txy' and shape (200, 130, 130).
Calculating Dominant frequencies and/or averaged power spectrum (Wavelet) ....
Progress: 100.00%
Analysis completed.
Processing Wavelet for a 3D cube with format 'txy' and shape (200, 130, 130).
Calculating Dominant frequencies and/or averaged power spectrum (Wavelet) ....
Progress: 100.00%
Analysis completed.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 | |
GPL Ghostscript 10.04.0 (2024-09-18)
Copyright (C) 2024 Artifex Software, Inc. All rights reserved.
This software is supplied under the GNU AGPLv3 and comes with NO WARRANTY:
see the file COPYING for details.
Processing pages 1 through 1.
Page 1
PDF saved in CMYK format as 'Figures/Fig4_dominant_frequency_mean_power_spectra.pdf'