Worked Example - NRMP: k-ω and POD Analysis¶
This example demonstrates the application of k-ω filtering and Proper Orthogonal Decomposition (POD) to a synthetic spatio-temporal dataset. The dataset consists of a time series of 2D images, representing the evolution of wave patterns over both space and time. By analysing this dataset with k-ω and POD, we can identify and isolate specific wave modes, revealing their spatial structures and temporal dynamics.
k-ω Analysis¶
k-ω analysis is a technique used to study wave phenomena in spatio-temporal datasets. It involves calculating the power spectrum of the data in both the spatial domain (wavenumber, k) and the temporal domain (frequency, ω). The resulting k-ω diagram shows how wave power is distributed across different spatial and temporal scales, providing insights into wave dispersion relations and the characteristics of different wave modes.
In this example, we apply k-ω analysis to the synthetic spatio-temporal dataset to identify and isolate a specific wave mode with a frequency of 500 mHz and wavenumbers between 0.05 and 0.25 pixel-1. We then use Fourier filtering to extract this wave mode from the dataset, revealing its spatial structure and temporal evolution.
Proper Orthogonal Decomposition (POD)¶
Proper Orthogonal Decomposition (POD) is a powerful technique for analysing multi-dimensional data. It identifies dominant spatial patterns, or modes, that capture the most significant variations in the data. POD is particularly useful for reducing the dimensionality of complex datasets and extracting coherent structures.
In this example, we apply POD to the synthetic spatio-temporal dataset to identify the dominant spatial modes of oscillation. We then apply frequency filtering to the temporal coefficients of these modes to isolate the 500 mHz wave mode. This allows us to compare the results of k-ω filtering and POD-based filtering, highlighting their respective strengths and limitations.
Analysis and Figure
The figure below shows a comparison of k-ω filtering and POD analysis applied to the synthetic spatio-temporal dataset.
Methods used:
- k-ω analysis with Fourier filtering
- Proper Orthogonal Decomposition (POD) with frequency filtering
WaLSAtools version: 1.0
These particular analyses generate the figure below (Figure 5 in Nature Reviews Methods Primers; copyrighted). For a full description of the datasets and the analyses performed, see the associated article. See the source code at the bottom of this page (or here on Github) for a complete analyses and the plotting routines used to generate this figure.
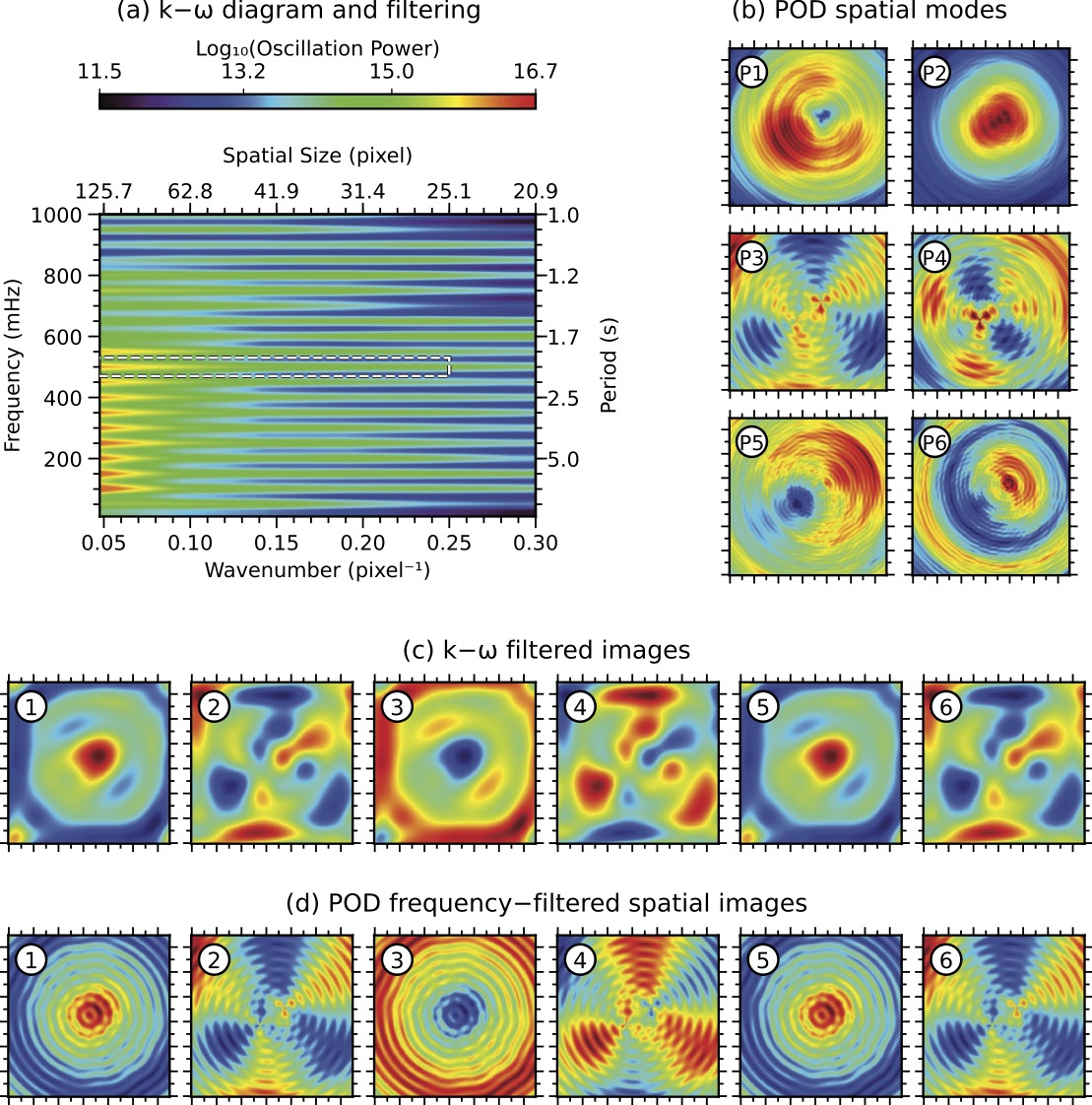
Figure Caption: Comparison of k-ω filtering and POD analysis. (a) k-ω power diagram of the synthetic spatio-temporal dataset with a targeted filtering region (dashed lines). (b) First six spatial modes from POD analysis (each 130×130 pixels2). © First six frames of the k-ω filtered datacube centred at 500 mHz (±30 mHz) and wavenumbers 0.05−0.25 pixel-1. (d) First six frames of the frequency-filtered POD reconstruction at 500 mHz using the first 22 POD modes (99% of total variance). All images and spatial modes are plotted with their own minimum and maximum values to highlight detailed structures within them.
Source code
© 2025 WaLSA Team - Shahin Jafarzadeh et al.
This notebook is part of the WaLSAtools package (v1.0), provided under the Apache License, Version 2.0.
You may use, modify, and distribute this notebook and its contents under the terms of the license.
Important Note on Figures: Figures generated using this notebook that are identical to or derivative of those published in:
Jafarzadeh, S., Jess, D. B., Stangalini, M. et al. 2025, Nature Reviews Methods Primers, 5, 21
are copyrighted by Nature Reviews Methods Primers. Any reuse of such figures requires explicit permission from the journal.
Free access to a view-only version: https://WaLSA.tools/nrmp
Supplementary Information: https://WaLSA.tools/nrmp-si
Figures that are newly created, modified, or unrelated to the published article may be used under the terms of the Apache License.
Disclaimer: This notebook and its code are provided "as is", without warranty of any kind, express or implied. Refer to the license for more details.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 | |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 | |
GPL Ghostscript 10.04.0 (2024-09-18)
Copyright (C) 2024 Artifex Software, Inc. All rights reserved.
This software is supplied under the GNU AGPLv3 and comes with NO WARRANTY:
see the file COPYING for details.
Processing pages 1 through 1.
Page 1
PDF saved in CMYK format as 'Figures/Fig5_k-omega_and_pod_analyses.pdf'