Worked Example - NRMP: POD Eigenvalues and Explained Variance¶
This example delves deeper into the analysis of Proper Orthogonal Decomposition (POD) results, focusing on the eigenvalues and explained variance. By examining the eigenvalues and their corresponding spatial modes, we can gain a better understanding of the dominant patterns and their contributions to the overall variability in the data.
Analysis and Figure
The figure below summarizes the POD analysis results, including the normalized eigenvalues, combined power spectrum, and cumulative explained variance.
Methods used:
- Proper Orthogonal Decomposition (POD)
- Power spectrum analysis
- Variance analysis
WaLSAtools version: 1.0
These particular analyses generate the figure below (Supplementary Figure S6 in Nature Reviews Methods Primers; copyrighted). For a full description of the datasets and the analyses performed, see the associated article. See the source code at the bottom of this page (or here on Github) for a complete analyses and the plotting routines used to generate this figure.
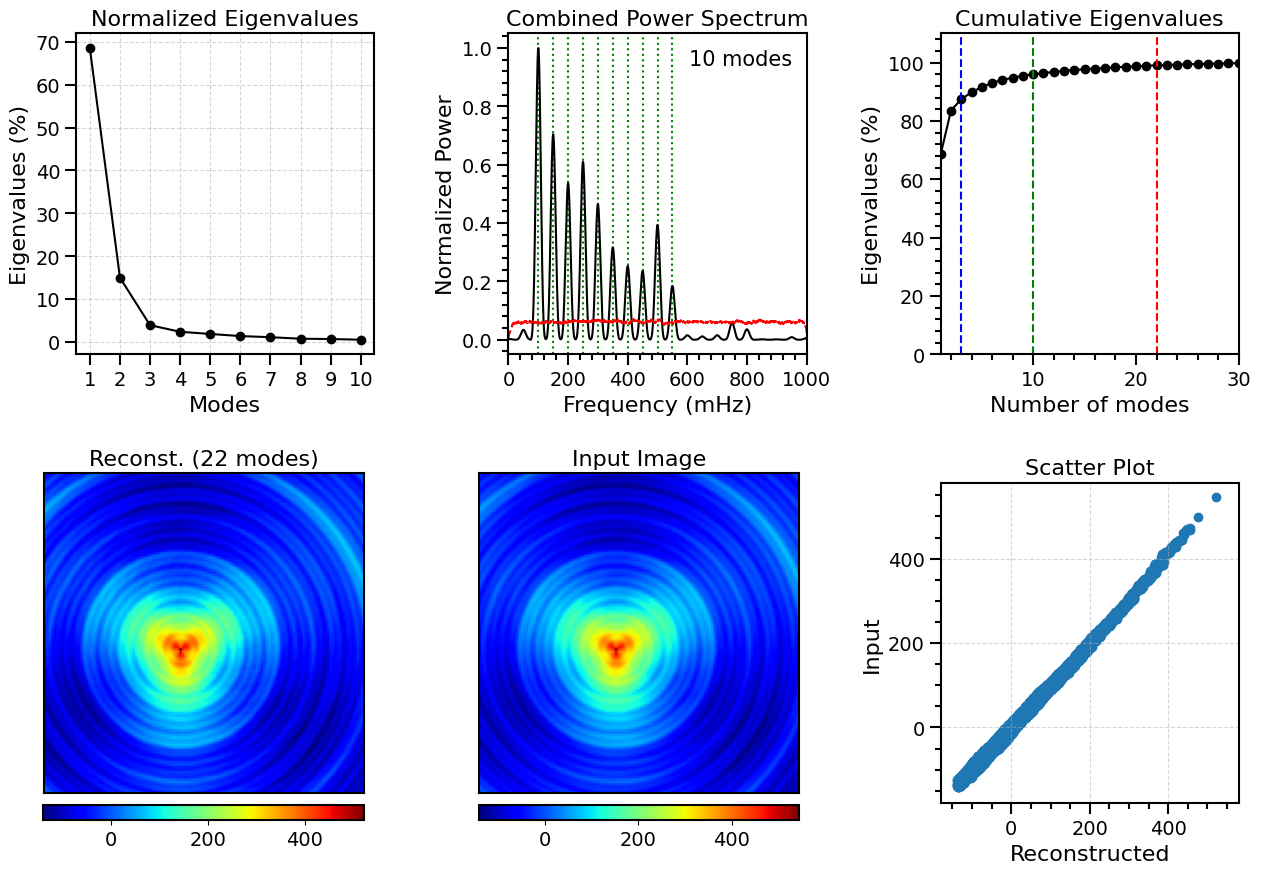
Figure Caption: POD mode analysis. Top left: Normalized squared singular values (eigenvalues) of the first ten POD modes, demonstrating their relative contributions to the total variance. Top middle: Combined power spectrum of the first ten POD modes, revealing the dominant frequencies captured by these ten modes. The vertical dotted lines mark the ten base frequencies used to construct the synthetic data; the red dashed line identifies the 95% confidence level (estimated from 1000 bootstrap resamples). Top right: Cumulative explained variance as a function of the number of POD modes included, with vertical lines indicating the cumulative variance captured by 2 (blue), 10 (green), and 22 (red) modes. Bottom left: Reconstructed image (130×130 pixels2) of the first frame of the time series using the first 22 modes. Bottom middle: Original image (130×130 pixels2; first frame) of the datacube. Bottom right: Scatter plot of the reconstructed and original image.
Source code
© 2025 WaLSA Team - Shahin Jafarzadeh et al.
This notebook is part of the WaLSAtools package (v1.0), provided under the Apache License, Version 2.0.
You may use, modify, and distribute this notebook and its contents under the terms of the license.
Important Note on Figures: Figures generated using this notebook that are identical to or derivative of those published in:
Jafarzadeh, S., Jess, D. B., Stangalini, M. et al. 2025, Nature Reviews Methods Primers, 5, 21
are copyrighted by Nature Reviews Methods Primers. Any reuse of such figures requires explicit permission from the journal.
Free access to a view-only version: https://WaLSA.tools/nrmp
Supplementary Information: https://WaLSA.tools/nrmp-si
Figures that are newly created, modified, or unrelated to the published article may be used under the terms of the Apache License.
Disclaimer: This notebook and its code are provided "as is", without warranty of any kind, express or implied. Refer to the license for more details.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 | |
Starting POD analysis ....
Processing a 3D cube with shape (200, 130, 130).
POD analysis completed.
Top 10 frequencies and normalized power values:
[[0.1, 1.0], [0.15, 0.7], [0.25, 0.61], [0.2, 0.54], [0.3, 0.47], [0.5, 0.39], [0.35, 0.32], [0.4, 0.25], [0.45, 0.24], [0.55, 0.18]]
Total variance contribution of the first 10 modes: 96.01%
---- POD/SPOD Results Summary ----
input_data (ndarray, Shape: (200, 130, 130)): Original input data, mean subtracted (Shape: (Nt, Ny, Nx))
spatial_mode (ndarray, Shape: (200, 130, 130)): Reshaped spatial modes matching the dimensions of the input data (Shape: (Nmodes, Ny, Nx))
temporal_coefficient (ndarray, Shape: (200, 200)): Temporal coefficients associated with each spatial mode (Shape: (Nmodes, Nt))
eigenvalue (ndarray, Shape: (200,)): Eigenvalues corresponding to singular values squared (Shape: (Nmodes))
eigenvalue_contribution (ndarray, Shape: (200,)): Eigenvalue contribution of each mode (Shape: (Nmodes))
cumulative_eigenvalues (list, Shape: (50,)): Cumulative percentage of eigenvalues for the first "num_cumulative_modes" modes (Shape: (num_cumulative_modes))
combined_welch_psd (ndarray, Shape: (8193,)): Combined Welch power spectral density for the temporal coefficients of the firts "num_modes" modes (Shape: (Nf))
frequencies (ndarray, Shape: (8193,)): Frequencies identified in the Welch spectrum (Shape: (Nf))
combined_welch_significance (ndarray, Shape: (8193,)): Significance threshold of the combined Welch spectrum (Shape: (Nf,))
reconstructed (ndarray, Shape: (130, 130)): Reconstructed frame at the specified timestep using the top "num_modes" modes (Shape: (Ny, Nx))
sorted_frequencies (ndarray, Shape: (21,)): Frequencies identified in the Welch combined power spectrum (Shape: (Nfrequencies))
frequency_filtered_modes (ndarray, Shape: (200, 130, 130, 10)): Frequency-filtered spatial POD modes for the first "num_top_frequencies" frequencies (Shape: (Nt, Ny, Nx, num_top_frequencies))
frequency_filtered_modes_frequencies (ndarray, Shape: (10,)): Frequencies corresponding to the frequency-filtered modes (Shape: (num_top_frequencies))
SPOD_spatial_modes (NoneType, Shape: None): SPOD spatial modes if SPOD is used (Shape: (Nspod_modes, Ny, Nx))
SPOD_temporal_coefficients (NoneType, Shape: None): SPOD temporal coefficients if SPOD is used (Shape: (Nspod_modes, Nt))
p (ndarray, Shape: (16900, 200)): Left singular vectors (spatial modes) from SVD (Shape: (Nx, Nmodes))
s (ndarray, Shape: (200,)): Singular values from SVD (Shape: (Nmodes))
a (ndarray, Shape: (200, 200)): Right singular vectors (temporal coefficients) from SVD (Shape: (Nmodes, Nt))
1 2 3 4 5 6 7 | |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 | |
GPL Ghostscript 10.04.0 (2024-09-18)
Copyright (C) 2024 Artifex Software, Inc. All rights reserved.
This software is supplied under the GNU AGPLv3 and comes with NO WARRANTY:
see the file COPYING for details.
Processing pages 1 through 1.
Page 1
PDF saved in CMYK format as 'Figures/FigS6_POD_egenvalues_powerspectrum.pdf'